TARGET ID/MOA
- How we identify the target protein of lead compound derived from phenotypic assay
-
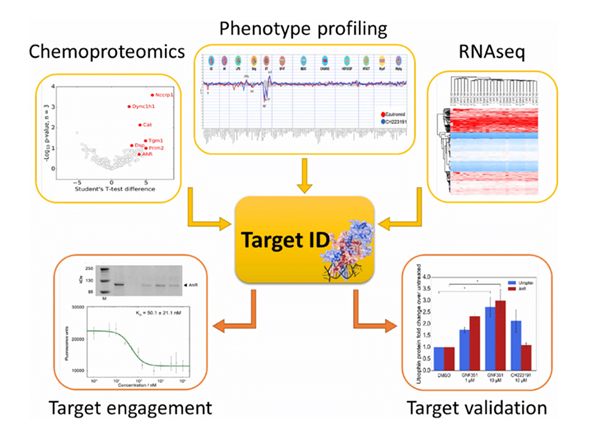
Phenotype-based drug discovery is an efficient strategy for identifying first-in-class drug candidates. Using phenotypic assay, unbiased discovery of bioactive hit compounds can be possible. Once hit compounds are identified, it is crucial to identify the target proteins of those compounds in order to understand the mechanism of action of those compounds and to advance to next development stages. However, the target identification and target validation are very challenging and laborious. That’s why many pharmaceutical companies cannot pursue phenotype-based drug discovery even though they know the high probability to identify novel hit compounds with new modes of action. Prof. Seung Bum PARK’s chemical biology laboratory at Seoul National University has been working on the development of new methods for target identification of bioactive compounds since 2010, and developed robust target deconvolution methods; photoaffinity-based FITGE (Fluorescence difference in Two-dimensional Gel Electrophoresis) method and label-free TS-FITGE (Thermal stability Shift-FITGE) method.
With these robust target deconvolution methods, SPARK Biopharma can reveal the target proteins of hit compounds and their incompletely understood mechanism of action (MOA). By exploring the early-stage target identification and the subsequent MOA study, SPARK Biopharma can facilitate phenotype-based drug discovery and bridge the phenotypic approach with rational drug discovery. The proteomics team at SPARK Biopharma has advanced analytical proteomics technology to provide information on broad biological contexts of targets and drug mechanisms on the basis of unbiased analysis of protein expression and functional analysis tools, which can provide an essential information for companion diagnosis.