FITGE/TS-FITGE
- How we utilize proteomics to discovery biomarkers
- The Phenotypic drug discovery is an efficient strategy for identifying promising small molecule drug candidates. In Phenotypic drug discovery, physiologically relevant biological system or cellular signaling pathway is directly interrogated by chemical compounds without bias. Once compounds are identified, it is crucial to identify the target proteins of those compounds in order to understand the mechanism of action of those compounds and to advance to next development stages. However, the target identification is very challenging and laborious. To accelerate the target identification of the compounds, we adopted the following technologies.
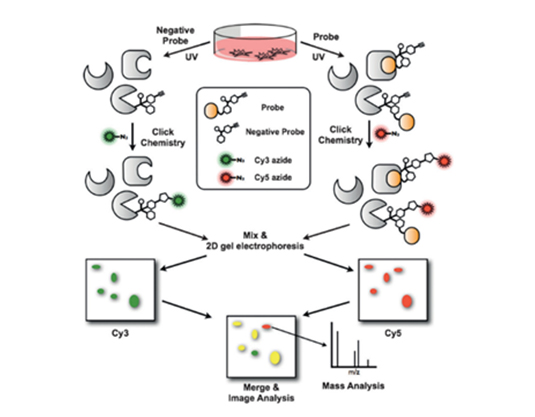
- FITGE (Fluorescence difference in Two-dimensional Gel Electrophoresis)
- FITGE is a robust target deconvolution method. The chemically attached reactive moiety enabled this bioactive molecule to bind to the target protein covalently. To visualize the drug-target complex, fluorescent moiety was introduced. Thus, the binding event can be monitored in the cells with intact signaling network. Then, the covalently modified proteins are identified after separation using 2D gel electrophoresis and LC-MS analysis. Until now, we have successfully identified target proteins of our compounds such as anticancer agent, antibacterial agent, and anti-inflammatory agent.
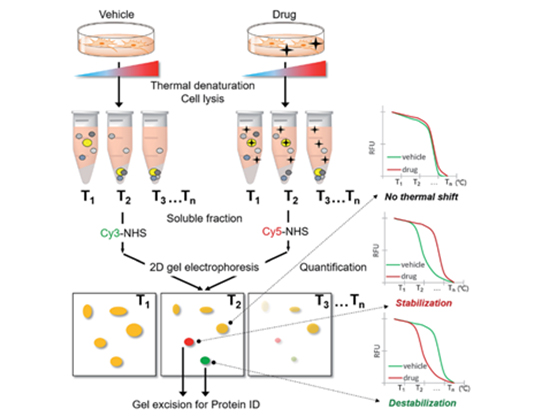
- TS-FITGE (Thermal stability Shift-FITGE).
- TS-FITGE, a modified version of FITGE, is a target deconvolution method without chemical modification of original bioactive molecules. TS-FITGE is based on a principle that protein-drug interaction increases thermal stability of target protein. The target protein was thermally stabilized upon drug binding, and we could differentiate the stabilized or destabilized target protein against all other proteins. TS-FITGE provides unbiased proteome-wide information on thermal stability shift by the interaction between compound and proteins.
Publications
-
1.
Phenotypic discovery of neuroprotective agents by regulation of Tau proteostasis via stress‐responsive activation of PERK signaling. Shin, Y.-H.; Cho, H.; Choi, B.Y.; Ha, J.; Kim, J.; Yim, J.; Suh, S.W.; Park, S.B.* Angew. Chem. Int. Ed., 2021, 60, 1831–1838
-
2.
Recent Advances in Identifying Protein Targets in Drug Discovery. Ha, J.†; Park, H.†; Park, J.*; Park, S.B.* Cell Chem. Biol. 2021, 28, 394–423.
-
3.
Label-free target identification reveals oxidative DNA damage as a mechanism of selective cytotoxic agent in cells with downregulated mismatch repair pathway. Park, H.; Park, S.B.* Chem. Sci., 2019, 10, 3449–3458.
-
4.
Label-Free Target Identification Using In-Gel Fluorescence Difference via Thermal Stability Shift. Park, H.; Ha, J.; Koo, J.Y.; Park, J.; Park, S.B.* Chem. Sci. 2017, 8, 1127–1133.
-
5.
Nonspecific Protein Labeling of Photoaffinity Linkers Correlates with Their Molecular Shapes in Living Cells. Park, H.†; Koo, J.Y.†; Srikanth, Y.V.V.; Lee, J.; Park, J.*; Park, S.B.* Chem. Comm. 2016, 52, 5828–5831.
-
6.
Investigation of Specific Binding Proteins to Photoaffinity Linkers for Efficient Deconvolution of Target Protein. Park, J.*; Koh, M.; Koo, J. Y.; Lee, S.; Park, S.B.* ACS Chemical Biology. 2016, 11(1), 44–52.
-
7.
A Small Molecule Binding HMGB1 and HMGB2 Inhibits Microglia-Mediated Neuroinflammation. Lee, S.; Nam, Y.; Koo, J.Y.; Lim, D.; Park, J.; Ock, J.; Suk, K.*; Park, S.B.* Nature Chemical Biology 2014, 10(12), 1055–1060.
-
8.
Phenotypic Screening to Identify Small-Molecule Enhancers for Glucose Uptake: Target Identification and Rational Optimization of Their Efficacy. Koh, M.; Park, J.; Koo, J.Y.; Lim, D.; Cha, M.Y.; Jo, A.; Choi, J.H.; Park, S. B.* Angew. Chem. Int. Ed. 2014, 53(20), 5102–5106.
-
9.
Discovery and Target Identification of an Antiproliferative Agent in Live Cells Using Fluorescence Difference in Two-Dimensional Gel Electrophoresis. Park, J.; Oh, S.; Park, S. B.* Angew. Chem. Int. Ed. 2012, 51(22), 5447–5451.
